Data storage handler¶
This data handler use PyTables that is built on top of the HDF5 library, using the Python language and the NumPy package. It features an object-oriented interface that, combined with C extensions for the performance-critical parts of the code (generated using Cython), makes it a fast, yet extremely easy to use tool for interactively browse, process and search very large amounts of data. One important feature of PyTables is that it optimizes memory and disk resources so that data takes much less space (specially if on-flight compression is used) than other solutions such as relational or object oriented databases.
[3]:
import tables
import numpy as np
import json
from datetime import datetime
[4]:
filename = 'sampple_eeg.h5'
CHANNELS = 16
[5]:
from openbci_stream.utils import HDF5Writer, HDF5Reader
Writer¶
[6]:
writer = HDF5Writer('output.h5')
Add header¶
[7]:
header = {'sample_rate': 1000,
'datetime': datetime.now().timestamp(),
}
writer.add_header(header)
Add EEG data¶
The number of channels is defined with first data addition:
[8]:
eeg = np.random.normal(size=(16, 1000)) # channels x data
writer.add_eeg(eeg)
The number of channels can not be changed after define the first package:
[9]:
eeg = np.random.normal(size=(8, 1000))
writer.add_eeg(eeg)
WARNING:OpenBCI-Stream:The number of channels 16 can not be changed!
Is possible to add any amount of data:
[10]:
eeg = np.random.normal(size=(16, 500))
writer.add_eeg(eeg)
A timestamp can be added to the data, in a separate command, in this case, timestamp could be an array of the same length of data.
[11]:
eeg = np.random.normal(size=(16, 2000))
timestamp = np.array([datetime.now().timestamp()]*2000)
writer.add_eeg(eeg)
writer.add_timestamp(timestamp)
Is possible to use the timestamp in a single command.
[12]:
eeg = np.random.normal(size=(16, 2000))
timestamp = [datetime.now().timestamp()] * 2000
writer.add_eeg(eeg, timestamp)
Extrapolated timestamps¶
Usually, we never have the datetimes for each value in a package, instead, we have a single marker time for each package. This module was designed keeping mind these cases, so, is possible to set a single timestamp and HDF5Reader will interpolate the missing timestamps
[13]:
eeg = np.random.normal(size=(16, 2000))
timestamp = datetime.now().timestamp()
writer.add_eeg(eeg, timestamp)
Add markers¶
The markers can be added individually with their respective timestamp:
[14]:
timestamp = datetime.now().timestamp()
marker = 'LEFT'
writer.add_marker(marker, timestamp)
Or in a dictionary structure with the marker as key and events as a list of timestamps:
[15]:
markers = {
'LEFT': [1603150888.752062, 1603150890.752062, 1603150892.752062],
'RIGHT': [1603150889.752062, 1603150891.752062, 1603150893.752062],
}
writer.add_markers(markers)
Add annotations¶
The annotations are defined like in EDF format, with an onset and duration in seconds, and their corresponding description:
[16]:
writer.add_annotation(onset=5, duration=1, description='This is an annotation, in EDF style')
writer.add_annotation(onset=45, duration=15, description='Electrode verification')
writer.add_annotation(onset=50, duration=3, description='Patient blinking')
Do no forget to close the file
[17]:
writer.close()
Reader¶
[27]:
reader = HDF5Reader('output.h5')
reader
[27]:
==================================================
output.h5
2021-04-10 20:17:21.274841
==================================================
MARKERS: ['LEFT', 'RIGHT']
SAMPLE_RATE: 1000
DATETIME: 1618103841.274841
SHAPE: [16, 7500]
==================================================
[20]:
reader.eeg
[20]:
array([[ 0.25868927, -0.66543329, 0.03229383, ..., 0.78450966,
0.574569 , -0.09854255],
[ 0.26966891, -0.09300477, -0.80611687, ..., -0.06674499,
-0.75905066, 0.69443338],
[-0.61651975, -0.98729215, -0.46595438, ..., -0.88060761,
-0.09198823, -0.81368015],
...,
[ 0.15696397, -0.05382042, -0.47447657, ..., -0.93922117,
-1.97117586, -1.97233181],
[-0.12692569, 0.17508998, 0.73853735, ..., 0.44395487,
0.88664937, 0.16267987],
[-0.86684959, -0.07770241, 0.30604455, ..., 2.16666748,
1.46009641, 1.20614713]])
[21]:
reader.markers
[21]:
{'LEFT': [1618103847.123672,
1603150888.752062,
1603150890.752062,
1603150892.752062],
'RIGHT': [1603150889.752062, 1603150891.752062, 1603150893.752062]}
[22]:
reader.f.root.eeg_data
[22]:
/eeg_data (EArray(16, 7500)) 'EEG time series'
atom := Float64Atom(shape=(), dflt=0.0)
maindim := 1
flavor := 'numpy'
byteorder := 'little'
chunkshape := (16, 512)
[23]:
reader.f.root.timestamp
[23]:
/timestamp (EArray(6000,)) 'EEG timestamp'
atom := Float64Atom(shape=(), dflt=0.0)
maindim := 0
flavor := 'numpy'
byteorder := 'little'
chunkshape := (8192,)
[24]:
reader.timestamp
[24]:
array([1.61810385e+09, 1.61810385e+09, 1.61810385e+09, ...,
1.61810385e+09, 1.61810385e+09, 1.61810385e+09])
[25]:
reader.annotations
[25]:
[[-1618103840.36316, 1, 'This is an annotation, in EDF style'],
[-1618103800.36316, 15, 'Electrode verification'],
[-1618103795.36316, 3, 'Patient blinking']]
[28]:
reader.close()
Examples using the with control-flow structure¶
This data format handlers can be used with the with structure:
Writer¶
This example create a file with random data:
[29]:
from openbci_stream.utils import HDF5Writer
from datetime import datetime, timedelta
import numpy as np
import os
now = datetime.now()
header = {'sample_rate': 1000,
'datetime': now.timestamp(),
'montage': 'standard_1020',
'channels': {i:ch for i, ch in enumerate('Fp1,Fp2,F7,Fz,F8,C3,Cz,C4,T5,P3,Pz,P4,T6,O1,Oz,O2'.split(','))},
}
filename = 'sample-eeg.h5'
if os.path.exists(filename):
os.remove(filename)
with HDF5Writer(filename) as writer:
writer.add_header(header)
for i in range(60*5):
eeg = np.random.normal(size=(16, 1000))
aux = np.random.normal(size=(3, 1000))
timestamp = (now + timedelta(seconds=i+1)).timestamp()
writer.add_eeg(eeg, timestamp)
writer.add_aux(aux)
# events = np.linspace(1, 59*30, 15*30).astype(int)*1000
# np.random.shuffle(events)
markers = {}
for i in range(5, 60*5//4, 4):
markers.setdefault('RIGHT', []).append((now + timedelta(seconds=i+1)).timestamp())
markers.setdefault('LEFT', []).append((now + timedelta(seconds=i+2)).timestamp())
markers.setdefault('UP', []).append((now + timedelta(seconds=i+3)).timestamp())
markers.setdefault('DOWN', []).append((now + timedelta(seconds=i+4)).timestamp())
writer.add_markers(markers)
writer.add_annotation((now + timedelta(seconds=10)).timestamp(), 1, 'Start run')
writer.add_annotation((now + timedelta(seconds=60)).timestamp(), 1, 'Head moved')
writer.add_annotation((now + timedelta(seconds=190)).timestamp(), 1, 'Blink')
Reader¶
[4]:
from openbci_stream.utils import HDF5Reader
from matplotlib import pyplot as plt
plt.figure(figsize=(16, 9), dpi=60)
ax = plt.subplot(111)
filename = 'sample-eeg.h5'
with HDF5Reader(filename) as reader:
channels = reader.header['channels']
sample_rate = reader.header['sample_rate']
t = np.linspace(0, 1, sample_rate)
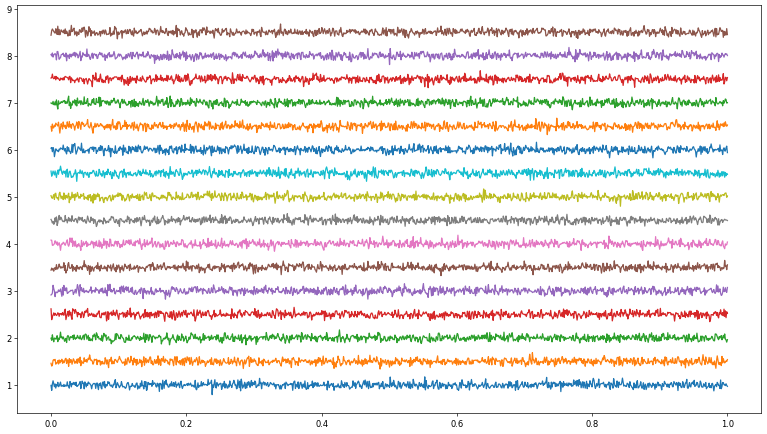
for i, ch in enumerate(reader.eeg[:, :sample_rate]):
plt.plot(t, (ch-ch.mean())*0.1+i)
ax.set_yticklabels(channels)
<ipython-input-4-ab8bda269688>:16: UserWarning: FixedFormatter should only be used together with FixedLocator
ax.set_yticklabels(channels)
MNE objects¶
reader.get_epochs() return an mne.EpochsArray object:
[31]:
reader = HDF5Reader('sample-eeg.h5')
epochs = reader.get_epochs(tmin=-2, duration=6, markers=['RIGHT', 'LEFT'])
mne.EpochsArray needs an array of shape (epochs, channels, time), we have channels and time, we only need to separate the epochs, here, an epoch is a trial, we will select trial by slicing around a marker. The argument duration in seconds set the width for the trials, and tmin set how much of signal before the marker will I analyze, finally, the argument markers set the labels to use, if empty, then will use
all of them.
This feature needs at least montage, channels, and sample_rate configured in the header.
[32]:
evoked = epochs['RIGHT'].average()
fig = plt.figure(figsize=(10, 5), dpi=90)
ax = plt.subplot(111)
evoked.plot(axes=ax, spatial_colors=True);
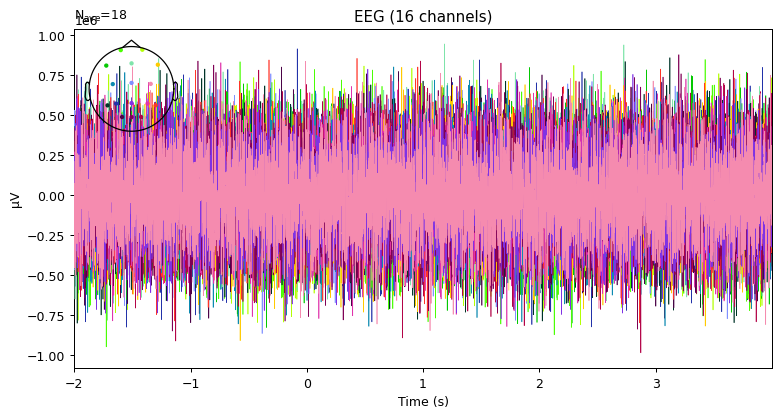
This is only random data.
[33]:
times = np.linspace(-1, 3, 6)
epochs['LEFT'].average().plot_topomap(times,)
epochs['RIGHT'].average().plot_topomap(times,);
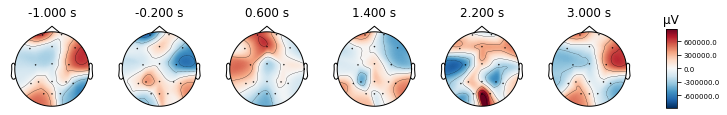
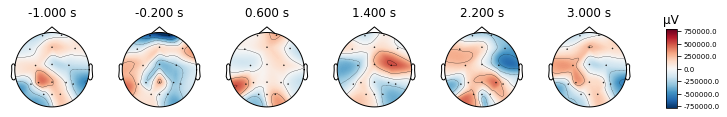
[16]:
reader.close()
Offset correction¶
The offset correction is performed through the start-offset and end-offset headers (created automatically), it corresponds to the respective offset at beginning and end on a real-time acquisition, this feature is enabled by default and can be changed with the argument offset_correction in the HDF5Reader class.
[ ]:
HDF5Reader('sample-eeg.h5', offset_correction=False)
RAW ndarray objects¶
get_data() return a standard array (epochs, channels, time) and their respective (classes), receives the same arguments that get_epochs:
[39]:
with HDF5Reader('sample-eeg.h5') as file:
data, classes = file.get_data(tmin=-2, duration=6, markers=['RIGHT', 'LEFT'])
data.shape, classes.shape
[39]:
((36, 16, 6000), (36,))
Export to EDF¶
The European Data Format (EDF) is a standard file format designed for exchange and storage of medical time series, is possible to export to this format with the to_edf method. This file format needs special keys in the header:
admincode: str with the admincode.
birthdate: date object with the the birthdate of the patient.
equipment: str thats describes the measurement equpipment.
gender: int with the the gender, 1 is male, 0 is female.
patientcode: str with the patient code.
patientname: str with the patient name.
patient_additional: str with the additional patient information.
recording_additional: str wit the additional recording information.
technician: str with the technicians name.
[36]:
reader = HDF5Reader('sample-eeg.h5')
reader.to_edf('sample-eeg.edf')
reader.close()
Relative time markers¶
In the previous examples, we are using the markers with a timestamp, this is an absolute reference, is an exact point in time, the reason of that is because this feature was designed for real-time streaming, in this kind of implementations the main reference is the system clock, this approach makes easy to implement a markers register in an isolated system.
But, is possible to get relative time markers, based on the position in the data array using the attribute reader.markers_relative